StructurE-unbiased Library for Exploratory disCovery of Targets
“From phenotype to pinpoint: decoding cancer drug targets with precision.”
“Unmasking viral vulnerabilities — where discovery meets mechanism.”
Antitumoral Target Deconvolution Pipeline
Uncovering the mechanisms of action behind chemotherapeutic agents.
The SELECT platform offers a proprietary 9-step pipeline to identify drug targets and mechanisms of action for bioactive compounds discovered through phenotypic screening. This pipeline integrates computational prediction, functional genetics, and chemical proteomics.
9-Step Pipeline Overview
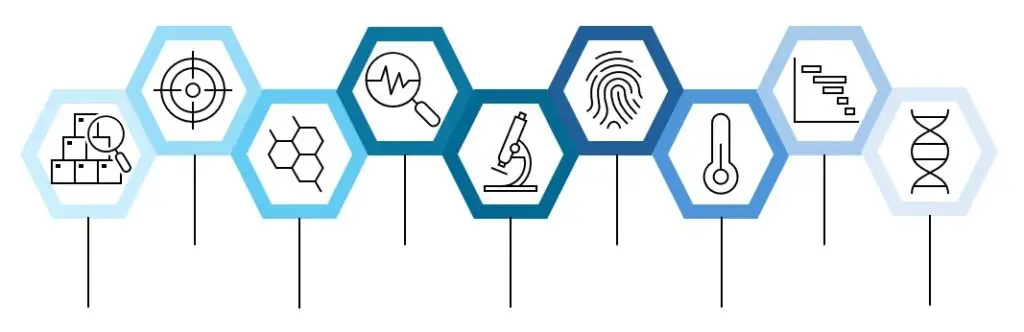
Computational Target Prediction
Common Mechanism Exclusion
Chemical Resynthesis & Validation
Toxicity Profiling: GDSC2 Comparison
Cell Painting
CRISPRres Mini Library Screen
Thermal Proteome Profiling (TPP)
CRISPRres Tiling Libraries
Genome-Wide CRISPR Screens
Mechanistic Insight Tools: CRISPRres, Morphological Profiling & Proteomics
CRISPRres Technology
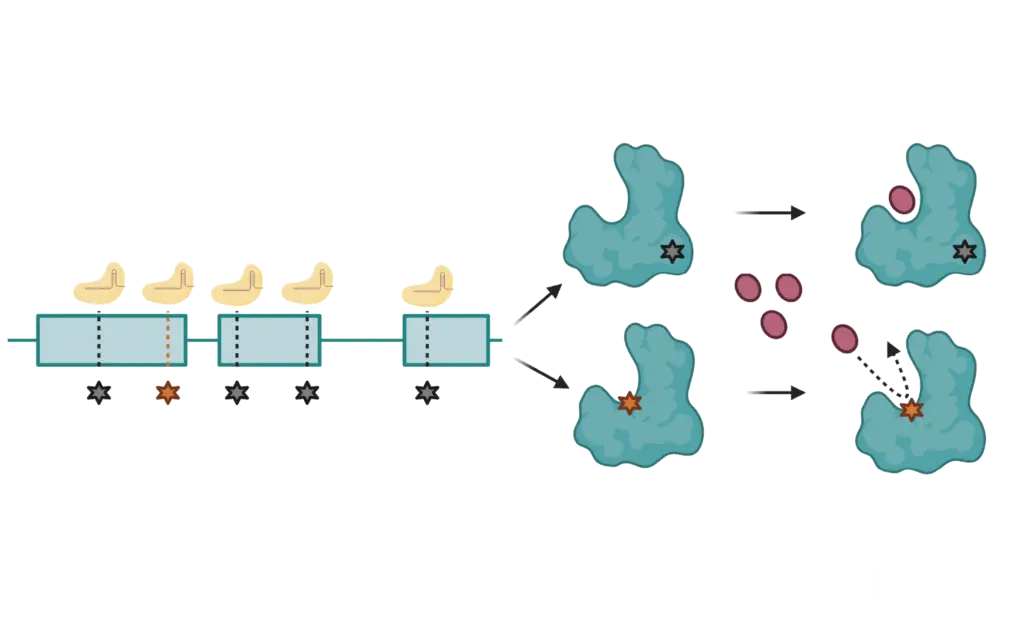
A patented CRISPR-based method (WO 2018/032063) that induces resistance mutations in essential genes to pinpoint drug targets and binding sites.
Cell Painting
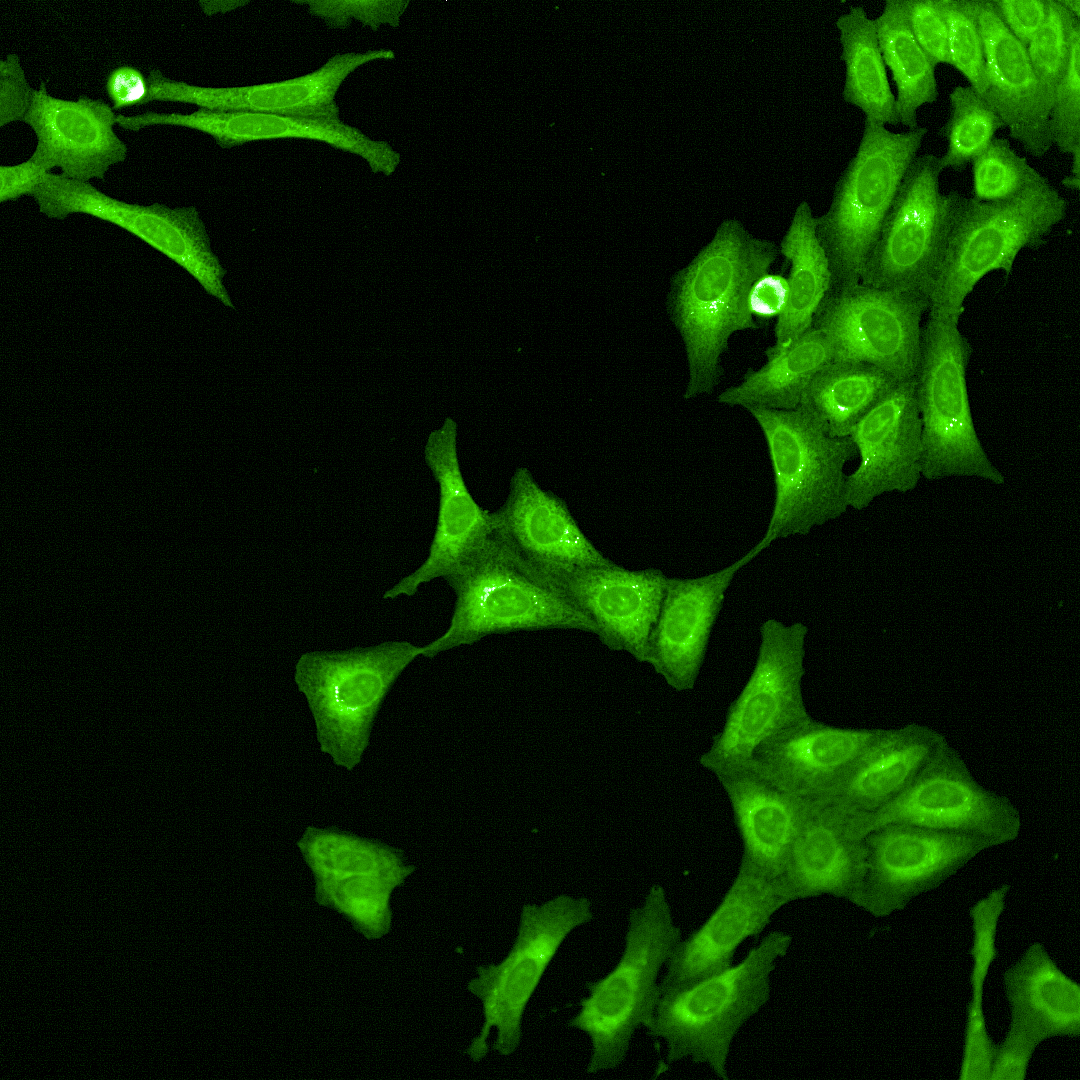
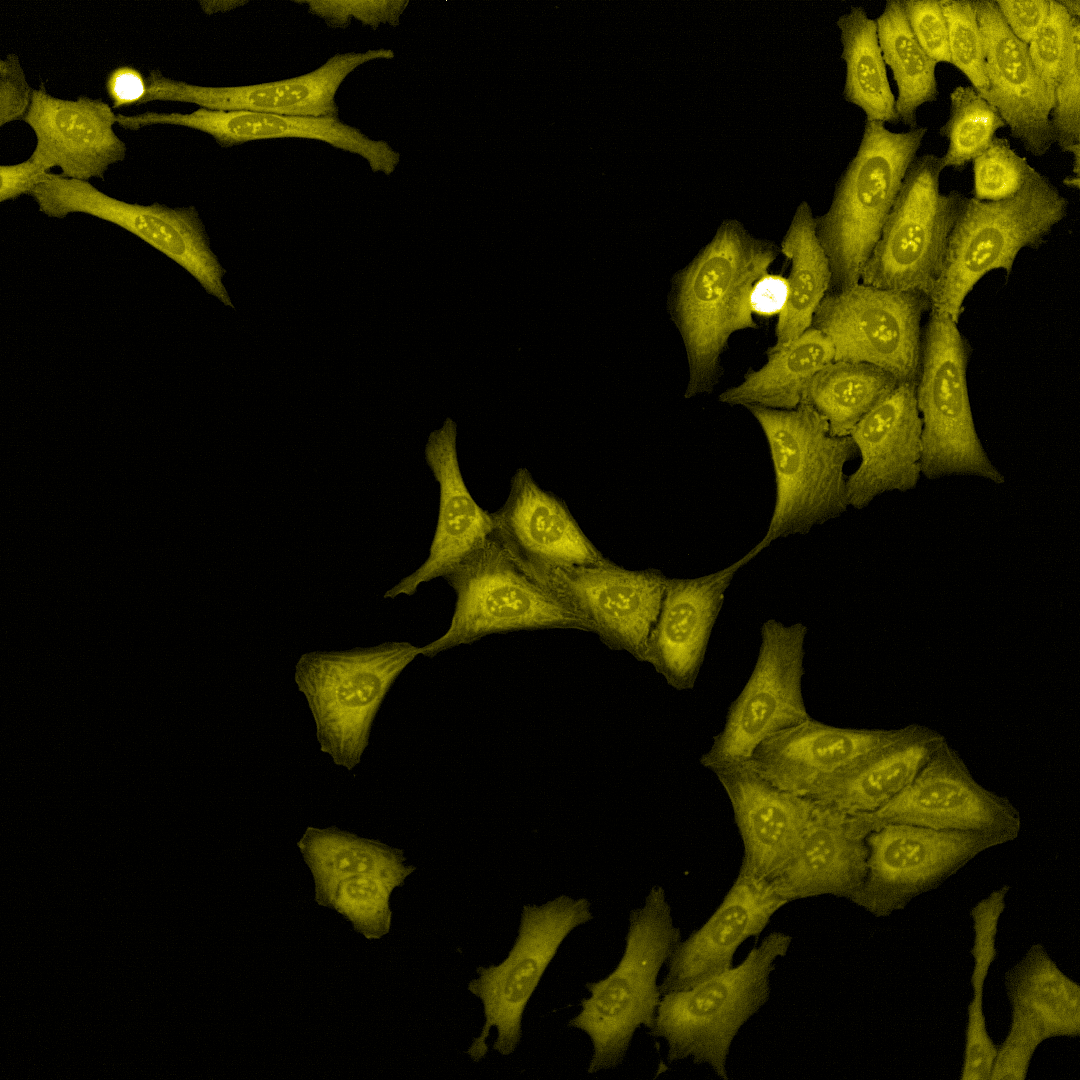
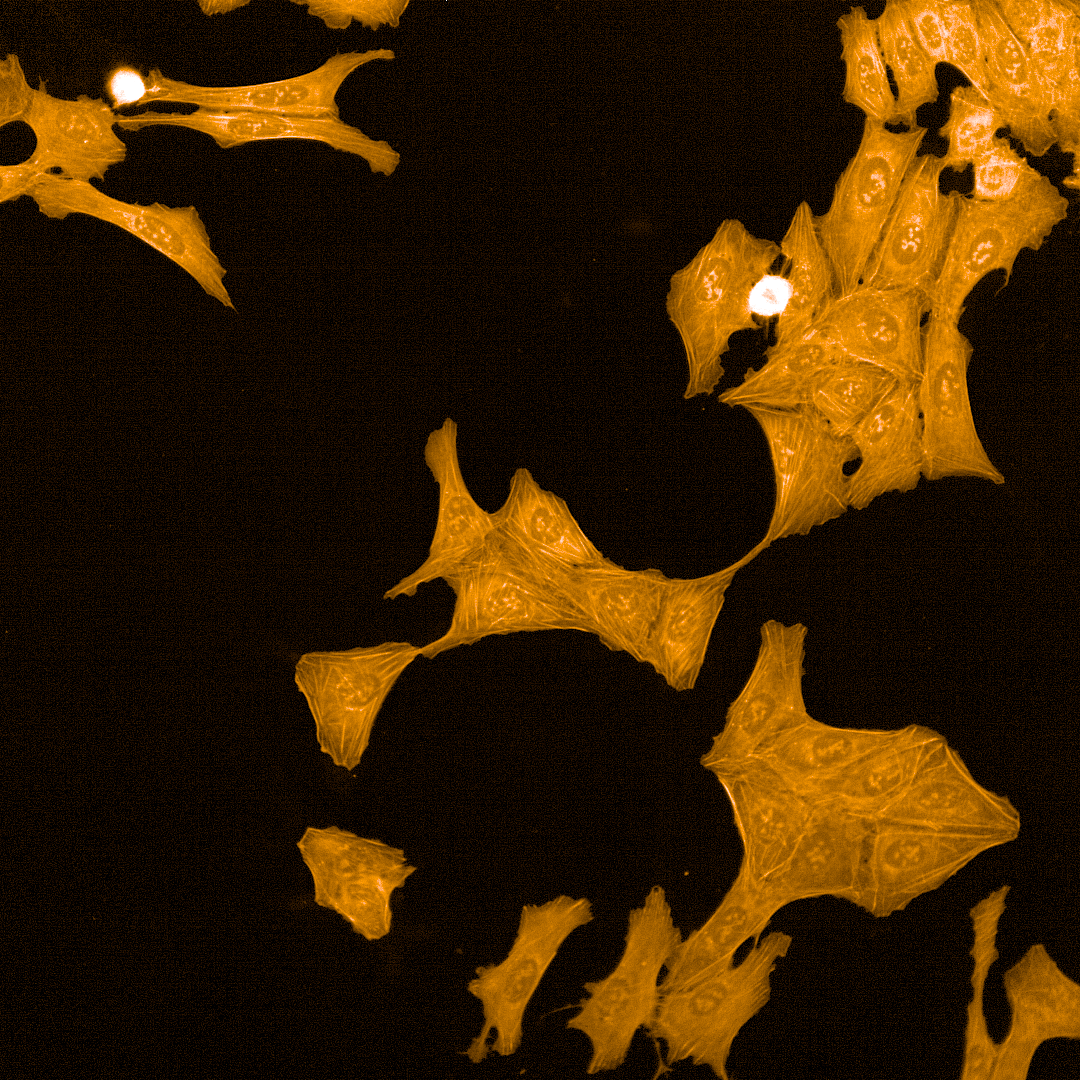
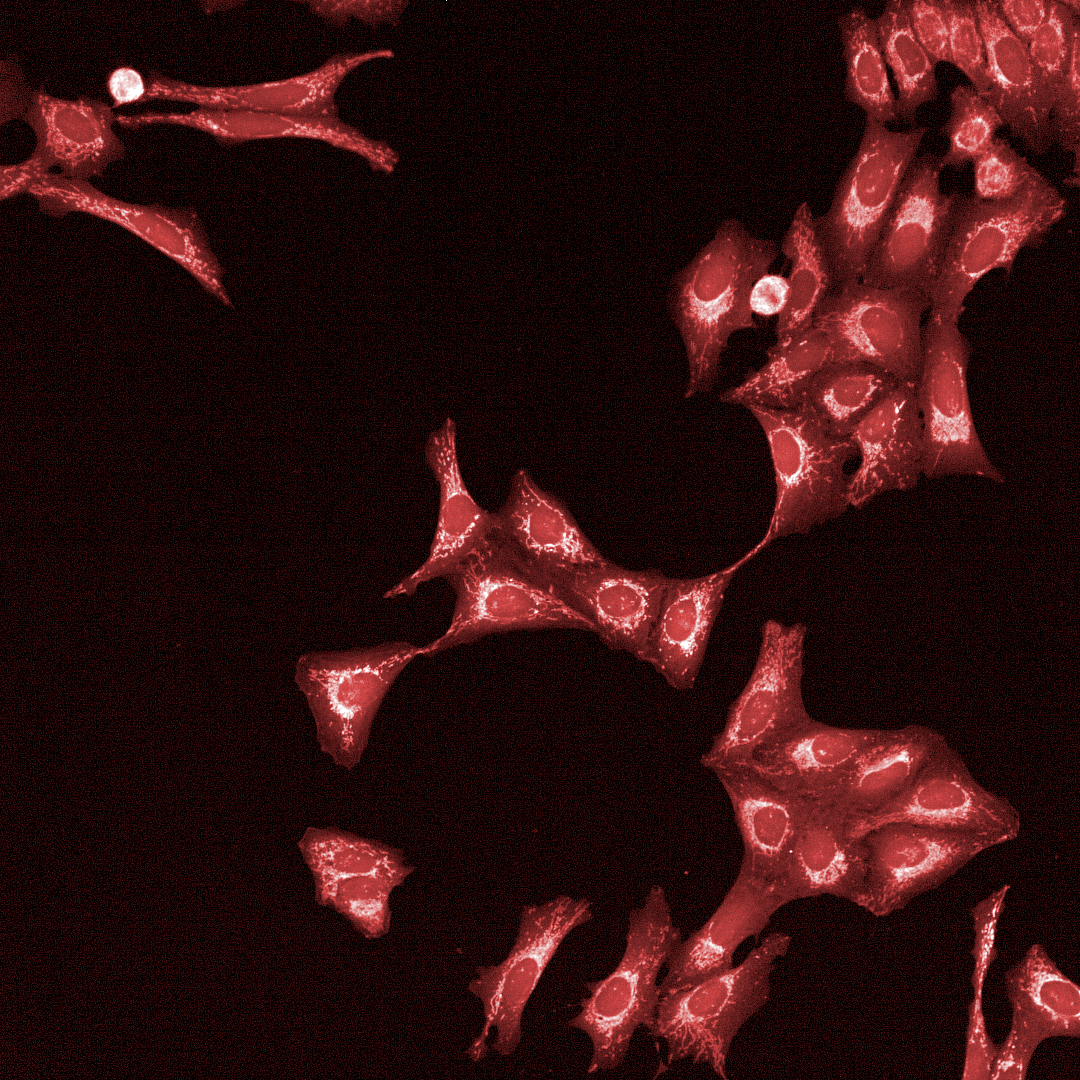
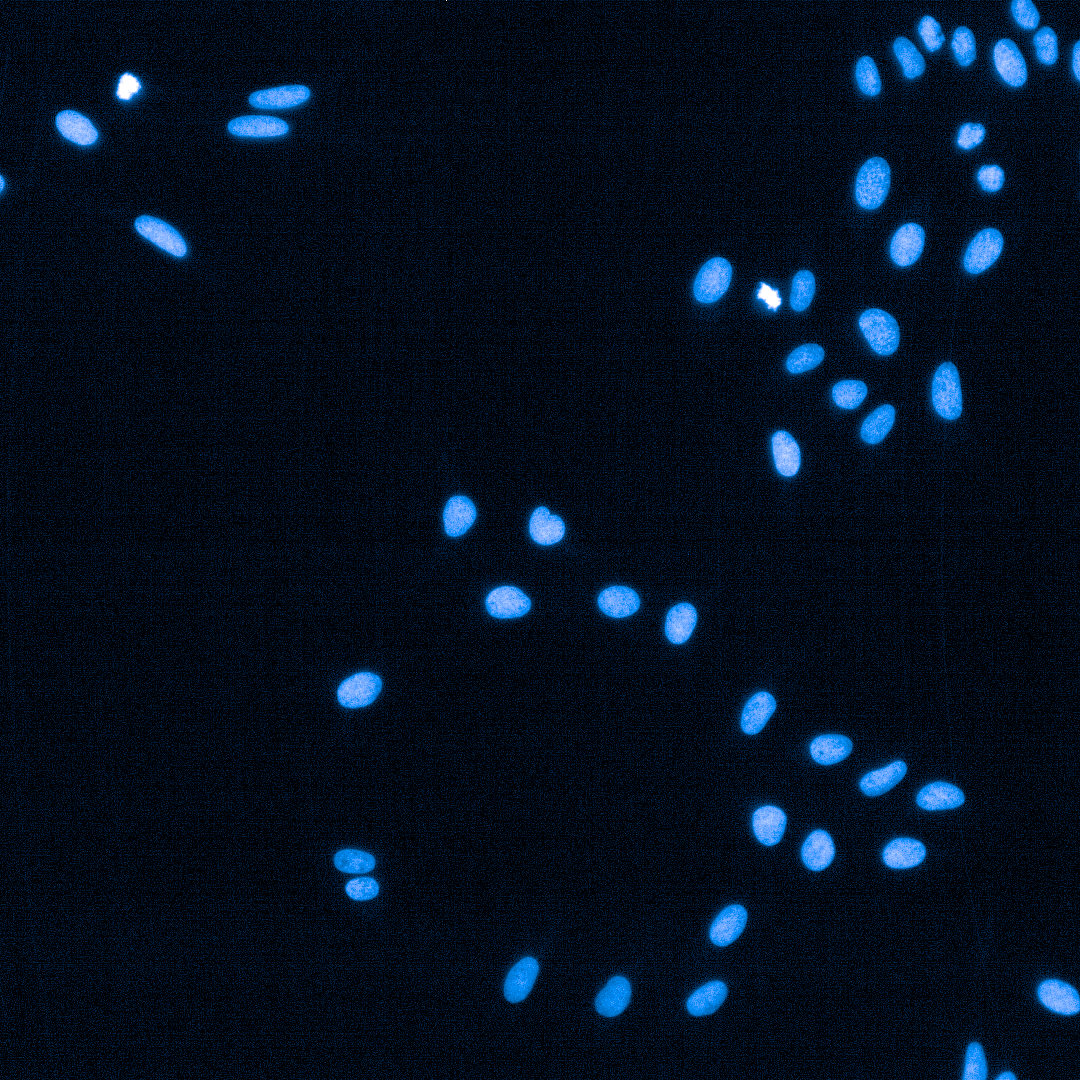
High-content imaging combined with AI-driven analysis to generate morphological fingerprints, revealing cellular pathways affected by compounds.
Proteomics
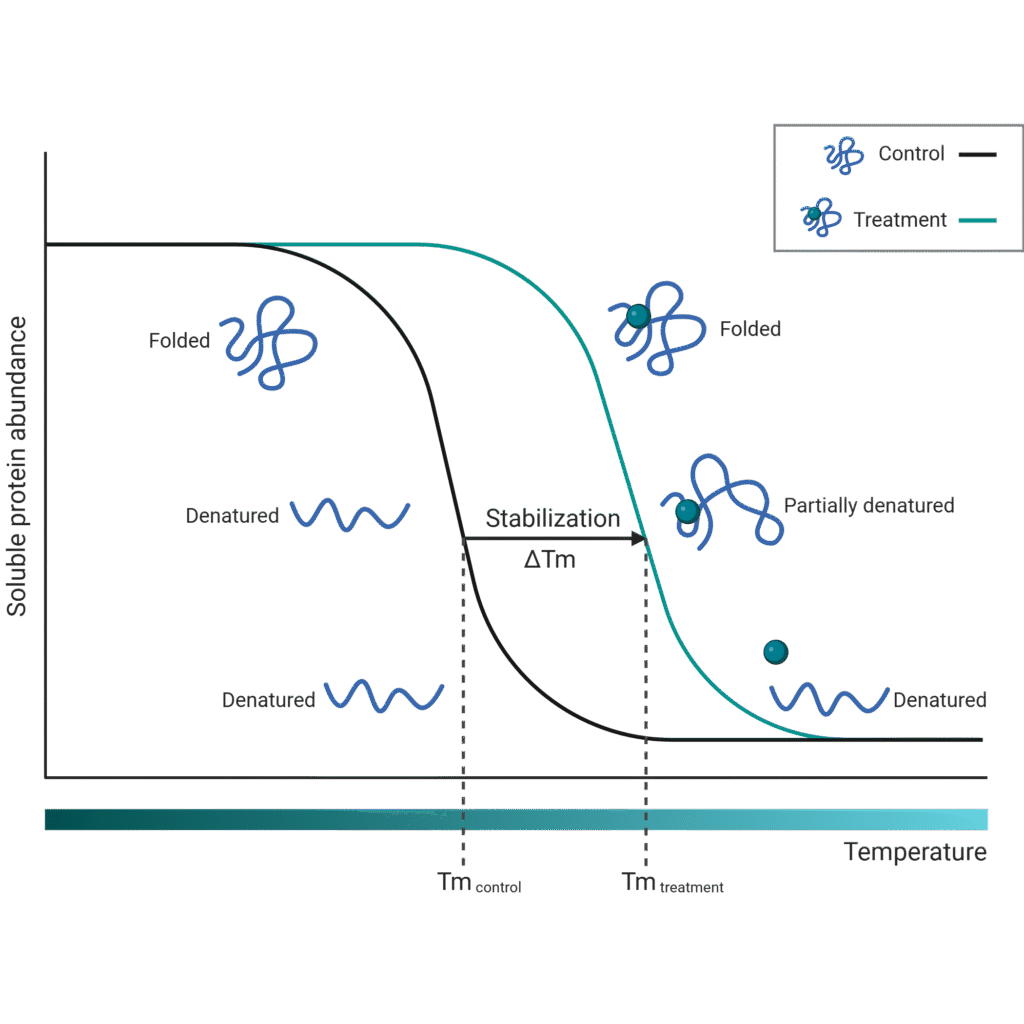
Thermal Proteome Profiling and related techniques to detect compound–protein interactions directly in live cells, uncovering mechanisms of action.
Learn More About Our Antitumoral Research
Dive deeper into the science behind our antitumoral target deconvolution pipeline; a unique, multi-step approach that combines computational prediction, functional genetics, and advanced proteomics to uncover mechanisms of action for promising compounds. This work enables the identification of novel drug targets and pathways that can transform cancer therapy. Explore ongoing projects, innovative methodologies, and collaborative opportunities by visiting the research group that drives this effort:
Antiviral Target Deconvolution
Elucidating the antiviral mechanism of bioactive molecules.
To support the mechanistic exploration of antiviral compounds, we employ a suite of in-house follow-up assays. These include cross-family viral screening to assess spectrum and selectivity, time-of-drug-addition experiments to pinpoint the stage of viral replication affected, fusion inhibition assays to evaluate interference with viral entry, and immunofluorescence-based imaging to visualize compound effects on viral protein expression and localization.
In parallel, we actively collaborate with specialized virology research teams within the Rega Institute, leveraging their deep expertise and advanced models for viruses such as flaviviruses, alphaviruses, retroviruses, and respiratory pathogens. This integrated approach ensures a robust and multidimensional characterization of antiviral hits emerging from the SELECT platform.